Phylogenetic relationships of Argentinean Creole horses and other South American and Spanish breeds inferred from mitochondrial DNA sequences
|
|
|
- Merilyn Stafford
- 5 years ago
- Views:
Transcription
1 Phylogenetic relationships of Argentinean Creole horses and other South American and Spanish breeds inferred from mitochondrial DNA sequences P. M. Mirol*, P. Peral García*, J. L. Vega-Pla and F. N. Dulout* *CIGEBA, Facultad de Ciencias Veterinarias, Universidad Nacional de La Plata, La Plata, Argentina. Laboratorio de Grupos Sanguíneos, Servicio de Cria Caballar, Córdoba, Spain Summary South American horses constitute a direct remnant of the Iberian horses brought to the New World by the Spanish conquerors. The source of the original horses was Spain, and it is generally assumed that the animals belonged to the Andalusian, Spanish Celtic, Barb or Arabian breeds. In order to establish the relationship between Argentinean and Spanish horses, a portion of the mitochondrial D-loop of 104 animals belonging to nine South American and Spanish breeds was analysed using SSCP and DNA sequencing. The variability found both within and between breeds was very high. There were 61 polymorphic positions, representing 16% of the total sequence obtained. The mean divergence between a pair of sequences was 2.8%. Argentinean Creole horses shared two haplotypes with the Peruvian Paso from Argentina, and the commonest haplotype of the Creole horses is identical to one of the Andalusian horses. Even when there was substantial subdivision between breeds with highly significant Wright s Fixation Index (FST), the parsimony and distance-based phylogenetic analyses failed to show monophyletic groups and there was no clear relationship in the trees between the South American and any of the other horses analysed. Although this result could be interpreted as mixed ancestry of the South American breeds with respect to the Spanish breeds, it is probably indicating the retention of very ancient maternal lineages in the breeds analysed. Keywords D-loop, horse, mtdna, phylogeny. Introduction The Argentinean Creole horse constitutes a direct remnant of the Iberian horses brought to the New World by the Spanish conquerors during the 15th century. The source of the original horses was Spain, and this was at a time when the Spanish horse was being used for improvement of horse breeding throughout Europe. On the basis of historical records, at least 250 horses where shipped to the continent, Address for correspondence P. M. Mirol, School of Biological Sciences, Queen Mary, University of London, Mile End Road, London E1 4NS, UK. p.m.mirol@qmul.ac.uk Accepted for publication 29 March 2002 from the second voyage of Columbus in 1493 to Nuñez de Cabeza s trip in 1540 (Rodero et al. 1992). These horses, as all other domestic animals transported, were quickly dispersed and became very well adapted to the new environment. At that time, Spanish exports arrived directly from the ports of Spain to their final destination, with a stop on the Canary Islands or the Antilles. There were in Spain three main morphological equine types: the Celtic type of tarpanic origin in the north and west region of the peninsula; the Spanish type, descendent from the African Barb horse, in the south and east; and finally, in the central area, the hybrid between both of them. As Seville, Cadiz and other southern ports monopolized the navigation to America, it could be assumed that horses taken to America were mainly of the Spanish type or Andalusian (Rodero et al. 1992).
2 MtDNA variation in South American and Spanish horses 357 However, the possibility of taking to America other equine types cannot be ruled out. During his second trip to the New World, Columbus complained to the Spanish Crown that the excellent horses shown to him before the departure had been changed by cheaper animals (Tudela 1987). In 1508 the Spanish Crown authorized the transport of 40 horses from Castilla in the expedition organized by Alonso Ojeda and Diego de Nicuesa to Panama. The horses were of the Celtic type, small and resistant. Furthermore, many animals died during the 2-month trip, and other animals from the intermediate ports, Canary Islands or Antilles, could have replaced them. The advent of mitochondrial DNA analysis in population genetics during the 1970s produced a revolutionary change regarding historical, biogeographic and phylogenetic perspectives on intra- and interspecific genetic structure (Avise 1994). Since then, it has been widely used to infer intra- and interspecific phylogenetic relationships. Mitochondrial DNA studies in horses have proved useful to characterize intrabreed variation (Kavar et al. 1999; Kim et al. 1999; Bowling et al. 2000), although retention of ancestral polymorphism makes phylogenetic inference difficult (Vilà et al. 2001). In this context we analyse the mitochondrial DNA variation of Argentinean Creole horses and some other South American and Spanish horses. The characterization of the Creole breed has been traditionally based on morphology and behaviour, and it was only during the last few years that analysis based on molecular genetic markers have been introduced (Peral García et al. 1996). Knowledge of the South American breeds is also important to conservation genetics of domestic horses, as the New World varieties are probably closer in type to the historical horse of Spain than are the current horses in Iberia, which over the last 500 years have interbred with other breeds. Materials and methods DNA extraction, PCR amplification and SSCP of the D-loop Total DNA was extracted from blood samples using the DNAZOL purification kit (Gibco Life Technologies, Rockville, MD, USA) following the manufacturer instructions. Samples included 45 Argentinean Creole horses (ARC), 30 Peruvian Paso from Argentina (PPA), 18 Arabian (16 from Argentina AR, and two from Spain ARSP) and 11 Spanish horses belonging to the breeds Asturcon (AST), Losino (LO), Potoka (PO), Mallorquina (MA), Menorquina (ME) and Spanish Pure Breed or Andalusian (AND). When pedigree information was available, we selected individuals that have not shared a common ancestor for at least three generations. The polymerase chain reaction (PCR) primers used were: L-strand: 5 -AGGACTATCAAAGGAGAAGCTCTA-3 (P1; Ishida et al. 1994) and H-strand: 5 -CCTGAAGTA- GGAACCAGATG-3 (H16498; Marklund et al. 1994), which amplify a 466-bp region situated between the trna Thr (position 15397; Xu & Árnason 1994) and the central domain of the D-loop (position 15863; Xu & Árnason 1994). The 50 ll reaction mix contained approximately 100 ng of total horse DNA, 0.5 lm of each primer, 0.1 mm of dntps and 2 U of Taq polymerase (Gibco BRL, Rockville, MD, USA) in 20 mm Tris HCl (ph 8.4), 50 mm KCl and 2mMMgCl 2, under mineral oil. The PCR consisted of a first denaturation step at 96 C for 2 min followed by 35 cycles of 1 min at 94 C, 30 s at 55 C and 1 min at 72 C, with an elongation step of 5 min at 72 C in the last cycle. The size of the products was estimated by 1.5% agarose gel electrophoresis with pbr322 MspI Digest as size marker. Argentinean Creole horses, Peruvian Paso from Argentina and Arabian (Argentina) were pre-selected before sequencing by PCR SSCP analysis, to include a range of genetically distinct individuals. Fifteen microliters of each PCR product was added to 40 ll of dye LIS (10% sucrose, 0.01% bromophenol blue and 0.01% xylene cyanol FF; Maruya et al. 1996). The samples were then heated at 96 C for 10 min, cooled on ice for at least 5 min and loaded onto a 10% polyacrylamide gel (49 : 1 acrylamide : bisacrylamide). Electrophoresis was carried out at 4 C, 200 V in 0.5 Tris-Borate-EDTA (TBE) buffer for 18 h. The gels were subsequently fixed in 5% ethanol, stained with 0.2% AgNO 3 and revealed with 2% CaCO 3. Cloning and sequencing of the PCR products The PCR products were purified using QIAquick columns (QIAGEN, Hilden, Germany) and cloned into dt-tailed pgem-t easy vector (Promega, Madison, WI, USA) following manufacturer s recommendations. The DNA sequencing was performed with an Applied Biosystems 377 automated sequencer (BioResource Centre, Cornell University, Ithaca, NY, USA) using T7 and M13 universal primers. At least two independent clones were sequenced from each individual and the unique substitutions were confirmed by sequencing clones from a second PCR product. Data analysis Sequences of the D-loop were aligned using CLUSTAL-V multiple alignment software (Higgins et al. 1992). Sites representing a gap in any of the aligned sequences were excluded from the analysis, and distances between D-loop sequences were estimated using both the absolute number of nucleotide differences and the Kimura two-parameter distance (Kimura 1980) calculated on the basis of an equal
3 358 Mirol et al. substitution rate per site. Phylogenies were constructed using maximum parsimony with the PAUP 4.0 software (Swofford 1997) and the NEIGHBOR program incorporated in the PHYLIP package (Felsenstein 1991). For this analysis sequences belonging to other horse breeds from GenBank ( were incorporated. In all analyses the sequence of Equus asinus (Xu & Arnason 1996) was used as outgroup. The statistical confidence of each node in the consensus trees was estimated by 1000 bootstrap resampling of the data. Analysis of molecular variance (AMOVA) and pairwise FST distances were calculated using Arlequin (Schneider et al. 2000). Results Single strand conformation polymorphism (SSCP) A total of 91 horses were examined for SSCP, 45 belonging to the Argentinean Creole breed, 16 Arabian and 30 Peruvian Paso. The PCR products were of the same length, approximately 460 base pairs, which is in agreement with the published horse sequences (Ishida et al. 1994). Heteroplasmy was not detected in any of the horses examined. The SSCP analysis revealed 14 variants (Table 1), which were consistently obtained in different runs and also in different SSCP conditions. As it can be seen from Table 1, Argentinean Creole and Peruvian Paso horses have five distinct SSCP patterns, and Arabian horses seven patterns. Arabian and Peruvian Paso present six and three diagnostic patterns, respectively (i.e. patterns not present in any other breed), while there are only two SSCP variants found exclusively in Argentine Creole horses. Three SSCP variants were shared amongst the breeds studied, two of them between Argentinean Creole and Peruvian Paso, and the other between Argentinean Creole and Arabian. The most common variants in each of the breeds analysed are unique to that specific breed. Sequence variation A total of 33 individuals from the Argentinean Creole, Peruvian Paso and Arabian breeds were sequenced, corresponding to the 14 SSCP variants found. At least one representative of each breed was sequenced for each one of the variants. When the resulting sequences were not identical for any particular SSCP pattern, all the individuals showing that particular variant were sequenced. Based on the analysis of 381 nucleotides between positions and 15827, 20 haplotypes were found. The 24 South American horses sequenced (ARC and PPA) showed 11 haplotypes. The number of haplotypes seems low compared with other breeds analysed [13 haplotypes in 16 maternal lines of Lipizzan horses (Kavar et al. 1999), 27 haplotypes in 34 Arabian maternal lines (Bowling et al. 2000)]. Although this result could be indicating a bottleneck during the establishment of the New World breeds, more information is needed to assess this phenomenon. Most haplotypes were restricted to a particular breed, but two of them where shared between Argentinean Creole and Peruvian Paso horses from Argentina. Three of the SSCP variants showed two different haplotypes each, with the extreme case of four different haplotypes showing an indistinguishable SSCP pattern (variant 3), differing in up to 15 nucleotides. Thirteen more individuals were sequenced from the Spanish breeds Asturcon, Losino, Potoka, Mallorquina, Menorquina, Spanish Pure breed or Andalusian and Arabian. All but one of these breeds generated new haplotypes. The only exception corresponded to one Andalusian animal (AND1), which shared its haplotype with Argentinean Creole horses (ARC1). This haplotype was the commonest found in Argentinean horses, corresponding to 26 animals showing SSCP variant 2, from which six horses were sequenced. They were not randomly selected, but rather were chosen by pedigree and also to represent three different breeders in the country. Analysis of the sequences showed 61 polymorphic positions, representing 16% of the total sequence obtained. The mean divergence between sequences was 2.8% (range %). All mutations detected corresponded to transitions, with one position (15534) representing an insertion/deletion of a single base pair located in the stretch of six cytosines. Number of animals showing each SSCP variant Table 1 Number and type of SSCP variants detected in each breed. Breed n nv ARC PPA AR ARC, Argentinean Creole; PPA, Peruvian Paso from Argentina; AR, Arabian, n ¼ number of individuals; nv ¼ number of SSCP variants.
4 MtDNA variation in South American and Spanish horses 359 Phylogenetic relationships Table 2 shows a summary of nucleotide diversities (p) and pairwise genetic distances among individuals within and between breeds. An AMOVA analysis showed substantial subdivision between breeds (FST ¼ , P < ), but with a large fraction of the variation (81.85%) found within populations. Furthermore, variation within breeds (x ¼ , SE ¼ ) was as high as variation between breeds (x ¼ , SE ¼ ). The data were first evaluated for phylogenetic information. The distribution of randomly generated trees was left-skewed (g 1 ¼ )0.6998, P < 0.01; Hillis & Huelsenbeck 1992), indicating phylogenetic signal in the data set. Maximum parsimony and neighbour-joining trees showed similar patterns. A heuristic search resulted in three equally parsimonious trees of length 123, 76 steps shorter than the shortest randomly generated tree. The consensus tree is shown in Fig. 1a. Two main clusters of haplotypes can be recognized. The first one includes haplotypes from Arabian (8 out of 11), Peruvian Paso (three out of six), Argentinean Creole (two out of seven), and all Menorquina and Potoka. The second group contains most of Argentinean Creole (five out of seven), half of the Peruvian Paso haplotypes, some Arabian (3 out of 11), Asturcon, one Mallorquina and all Andalusian and Losino. The neighbourjoining tree (Fig. 1b) differs in the clustering of AND3, ARC2, ARB3, PPA4, LO1 and LO2, which in the distance analysis appeared as a clade related to the first group of haplotypes. Although the results of a bootstrap analysis with 1000 replications showed significant probabilities for most of the external nodes, only the second group of sequences resulted in a significant bootstrap value (79%), which is mainly because of shared substitutions at positions 15494, 15496, 15534, and The lack of support of most of the nodes is not surprising given the previous bootstrap values found in most of the phylogenetic analysis of horses published to date, where only a few nodes of the reconstructed phylogeny showed significant probabilities (Kavar et al. 1999; Kim et al. 1999; Bowling et al. 2000). Both maximum parsimony and neighbour-joining analysis showed no clear relationship between the South American breeds (Argentinean Creole and Peruvian Paso) and any of the Spanish breeds analysed, apart from the decisive fact of Argentinean Creole and Andalusian horses sharing one haplotype. In order to compare the results obtained using mitochondrial DNA and previous results based on microsatellite data (Cañon et al. 2000), maximum parsimony and neighbour-joining trees were also constructed for the subset of Spanish horses (trees not shown). Parsimony and distance analysis resulted in identical trees, with bootstrap values higher than 50% in most of the nodes. There was a first cluster of sequences including ARSP, Potoka and Menorquina breeds (bootstrap 51%). Each one of the three breeds represented in this group was monophyletic with bootstrap values higher than 90%. A second group consisted of Losino and one Andalusian haplotype (bootstrap 78%), and a third group included AST, AND1 and MA2 (bootstrap 86%). As in the trees in Fig. 1, the MA1 haplotype did not cluster with any other sequence. The results are quite different from the ones obtained using microsatellites (Cañon et al. 2000), where a clear clustering of the Atlantic breeds (including Asturcon, Potoka, and Losino) vs. a cluster of Mediterranean breeds (Mallorquina and Menorquina) was obtained. We also analyse the sequences reported in this work in a wider context. Vilà et al. (2001) have found that a wide variety of mitochondrial haplotypes of horse breeds clustered in seven different clades with low bootstrap support, indicating a high number of ancestral matrilines of ancient origin. We selected 19 haplotypes (accession numbers: AF326677, AF326676, AF326678, AF356672, Table 2 Nucleotide diversity (p) and mean number and range of nucleotide differences within and between breeds. References as in Table 1. Intra-breed values are in bold. ARC PPA AR AND AST LO PO MA ME p ARC 6.92 (1 13) 8.71 (0 15) (3 19) 7.21 (0 11) 8.71 (4 13) 9.57 (7 13) (6 18) 7.71 (1 13) (9 16) PPA 7.77 (1 15) 11 (4 20) 8.5 (1 14) 11 (5 14) 8.75 (6 13) 11.5 (5 19) 9.12 (3 14) 10.5 (5 17) AR (2 20) (2 17) (6 21) (5 15) (1 17) (2 18) (3 17) AND 10 7 (4 10) 6.5 (2 12) 14 (13 15) 7.5 (2 10) 11.5 (10 13) AST 10 (8 12) 18 (17 19) 9 (6 12) 13.5 (12 15) LO 4 12 (11 13) 9.5 (6 12) 10.5 (8 13) PO 2 14 (11 17) 11.5 (9 14) MA (10 15) ME 3
5 360 Mirol et al. (a) Figure 1 (a) Consensus of the three most parsimonious trees found in the sample of South American and Spanish horses analysed. Figures on the internodes are bootstrap probabilities (in percentage) based on 1000 replications. (b) Neighbour-joining tree based on the Kimura two-parameters distances. Bootstrap values on the internodes. Haplotype names as in the text. AF326674, AF326669, AF064628, AF014416, AF014410, AF072996, AF14413, AF072990, AF072988, AF072992, AF072987, AF014409, AF064632, AF072977, D23666) in order to represent the seven clades described. A maximum parsimony and neighbour-joining analysis were conducted with the total data set of 53 sequences. The neighbour-joining tree is shown in Fig. 2. The structure of the tree is the same as the one obtained in Vilà et al. (2001). The Spanish and South American haplotypes are dispersed along the different clades. Discussion In a previous analysis based on five polymorphic protein loci, Peral García et al. (1996) found a close relationship between Andalusian, Barb, Argentinean Creole horses and Peruvian Paso. Bowling (1994) has also suggested a close association between Andalusian and Argentinean Creole based on blood type markers, although in this case the Nei s distances among all breeds and feral populations examined were so similar that the results were not conclusive. Our results indicate that there is a close relationship between Andalusian and Argentinean Creole horses. It is very noteworthy that ARC1, the most common haplotype found in the South American breed with 26 animals showing SSCP variant 2, is identical to one of the haplotypes found in the Andalusian horses examined. A longer sequence of 468 bp (positions ) was compared between both haplotypes and proved to be identical.
6 MtDNA variation in South American and Spanish horses 361 (b) Figure 1b Continued. The mean number of nucleotide differences between both breeds was 7.21 (range 0 11), the lowest of all interbreed comparisons, and very similar to the mean number of nucleotide differences within the Argentinean Creole horses (6.92, range 1 13). The Nei s genetic distances were higher in all comparisons with Celtic and Arabian horses. With the Peruvian Paso from Argentina, the mean number of nucleotide differences was 8.71 (range 0 15). There were two haplotypes shared between both breeds (CP1 and CP2). The origin of the Peruvian Paso can be traced to the 16th century, from Barb and Andalusian horses brought by Spanish conquerors to Peru, so their close association with the Argentinean breed is not surprising. Furthermore, the Argentinean Peruvian Paso is a young breed, and so, even when stallions were brought from Peruvian Paso in Peru, the mares were frequently not pure Peruvian Paso, and their mtdna was probably of mixed ancestry. The mean number of nucleotide differences between Peruvian Paso and Andalusian is 8.50 (range 1 14), again the lowest of all comparisons and similar to the comparison Argentinean Creole Peruvian Paso and the Peruvian Paso intrabreed differences (7.78, range 1 15). Even when the distances between Argentinean Creole, Peruvian Paso and Andalusian are low, haplotypes representing both Argentinean Creole and Peruvian Paso appeared in the two main clades found in the parsimony and neighbour-joining trees (Fig. 1a,b). This could be interpreted as mixed ancestry or multiple origin of the South American breeds, as was suggested when similar results were obtained in studies of mtdna of Lipizzan horses (Kavar et al. 1999) and Cheju horses (Kim et al. 1999). An alternative explanation could be that the phylogenetic
7 362 Mirol et al. Figure 2 Parsimony tree obtained when some of the sequences reported in Vilà et al. (2001) were included in the analysis. Pleist: remains of horses dated years ago, Anc: remains of horses dated years ago (Vilà et al. 2001). Figures in the internodes represent bootstrap values, and letters indicate the different clades described by Vilà et al. In bold and underlined, sequences from the horse breeds analysed in the present work. reconstruction is reflecting very ancient maternal lineages, present in the Spanish ancestors and so in the South American descendants. This hypothesis is in agreement with the tree depicted in Fig. 2. Haplotypes of Argentinean Creole horses appeared in four different clades (A, C, D and F, nomenclature of Vilà et al. 2001), and Argentinean Peruvian Paso in three clades (A, C and D). Andalusian horses are clustered in clades C and D, which also contain Argentinean Creole and Peruvian Paso. Vilà et al. (2001) propose that modern horse sequences do not define monophyletic groups with respect to wild progenitors, as would be expected if they were founded from a limited wild stock. They assume that the high diversity of matrilines observed suggests the use of a large number of wild populations in the origin of the domestic horse. In this context, the lack of strongly supported phylogenetic relationships among the breeds analysed in this work, indicates the retention of very ancient mitochondrial diversity. The phylogenetic pattern is rather different if microsatellites are used as molecular markers. As the development of distinct breeds usually follows a pattern of very restricted selection based in a few males serving many females, individuals from the same breed are generally clustered together in a phylogenetic tree when microsatellites are used (Vilà et al. 2001). In a previous work, Cañon et al. (2000) studied the genetic structure of Spanish Celtic horses, including Asturcon, Losino, Potoka, Mallorquina and Menorquina, using microsatellites. The genetic distances between breeds
8 MtDNA variation in South American and Spanish horses 363 were in all cases higher than the ones we obtained here using mtdna (Table 2), and many of the breeds were defined as monophyletic groups. They also found a clear clustering among all Atlantic breeds (Asturcon, Losino, Potoka), different from the clade containing the Mediterranean breeds (Mallorquina and Menorquina). This tree is different from the one based on mtdna, where only two of the Spanish breeds (Potoka and Menorquina) constitute monophyletic groups, and there is no association between Atlantic breeds on the one hand, and Mediterranean breeds on the other. This difference is reflecting the maternally dominated genetic flow between breeds and the male-biased selection in the breeds development. In conclusion, we provide the first mitochondrial characterization of South American and Spanish horse breeds. In addition, our results support the very ancient origin of the matrilines in horses, from the perspective of modern New World breeds. Accession numbers GenBank accession numbers for the sequences presented here are AF to AF Acknowledgements This paper was supported by National Research Council and Universidad Nacional de La Plata grants to PPG and FND, and SECyT and Fundación Antorchas grants to PMM. The authors wish to thank Mariana Kienast for the collection of the samples and Jeremy B. Searle and Chris Faulkes for very useful comments. References Avise J.C. (1994) Molecular Markers, Natural History and Evolution. Chapman & Hall, New York. Bowling A.T. (1994) Population genetics of Great Basin feral horses. Animal Genetics 25, Bowling A.T., Del Valle A. & Bowling M. (2000) A pedigree-based study of mitochondrial D-loop sequence variation among Arabian horses. Animal Genetics 31, 1 7. Cañon J., Checa M.L., Carleos C., Vega Pla J.L. & Dunner S. (2000) The genetic structure of Spanish Celtic horse breeds from microsatellite data. Animal Genetics 31, Felsenstein (1991) PHYLIP (Phylogeny Inference Package), Version 3.4. University of Washington, Seattle, WA. Higgins D.G., Bleasby A.J. & Funchs R. (1992) CLUSTAL V: improved software for multiple sequence alignment. Computer Applications in the Biosciences 8, Hillis D.M. & Huelsenbeck J.P. (1992) Signal, noise and reliability in molecular phylogenetic analysis. Journal of Heredity 83, Ishida N., Hasegawa T., Takeda K., Sakagami M., Onishi A., Inumaru S., Kamtsu M. & Mukoyama H. (1994) Polymorphic sequence in the D-loop region of equine mitochondrial DNA. Animal Genetics 25, Kavar T., Habe F., Brem G. & Dovc, P. (1999) Mitochondrial D-loop sequence variation among the 16 maternal lines of the Lipizzan horse breed. Animal Genetics 30, Kim K.-I., Yang Y.-H., Lee S.-S., Park C., Ma R., Bouzat J.L. & Lewin H.A. (1999) Phylogenetic relationships of Cheju horses to other horses breeds as determined by mtdna D-loop sequence polymorphism. Animal Genetics 30, Kimura M. (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16, Marklund S., Chaudhary R., Marklund L., Sandberg K. & Andersson L. (1994) Extensive mtdna diversity in horses revealed by PCR SSCP analysis. Animal Genetics 26, Maruya E., Saji H. & Yokoyama S. (1996) PCR LIS SSCP (Low ionic strenght stranded conformatioon polymorphism) a simple method for high resolution allele typing of HLA-DQB1, and DPB1. Genome Research 6, Peral García P., Kienast M., Villegas E., Díaz S. & Dulout F. (1996) Estudio de relaciones genéticas entre razas equinas mediante el análisis multivariado. Agro Sur 24 (1), Rodero, A., Delgado, J.V. & Rodero, E. (1992) Primitive Andalusian livestock and their implications in the discovery of America. Archivos de Zootecnia 41, Schneider S., Roessli D. & Excoffier L. (2000) Arlequin: A Software for Population Genetics Data Analysis, Ver Genetics and Biometry Laboratory, Department of Anthropology, University of Geneva, Geneva. Swofford D.L. (1997) PAUP: Phylogenetic Analysis Using Parsimony. Smithsonian Institution, Washington, DC. Tudela J. (1987) El Legado de España a America. Ediciones Pegaso, Madrid. Vilà C., Leonard J.A., Götherström A., Marklund S., Sandberg K., Lidén K., Wayne R.K. & Ellegren H. (2001) Widespread origins of domestic horse lineages. Science 291, Xu X. & Árnason U. (1994) The complete mitochondrial DNA sequence of the horse, Equus caballus: extensive heteropplasmy of the control region. Gene 148, Xu X. & Árnason U. (1996) The complete mitochondrial DNA (mtdna) of the donkey and mtdna comparison among four closely related mammalian species-pairs. Journal of Molecular Evolution 43,
Mitochondrial DNA D-loop sequence variation among 5 maternal lines of the Zemaitukai horse breed
 Research Article Genetics and Molecular Biology, 28, 4, 677-681 (2005) Copyright by the Brazilian Society of Genetics. Printed in Brazil www.sbg.org.br Mitochondrial DNA D-loop sequence variation among
Research Article Genetics and Molecular Biology, 28, 4, 677-681 (2005) Copyright by the Brazilian Society of Genetics. Printed in Brazil www.sbg.org.br Mitochondrial DNA D-loop sequence variation among
Genetic Relationship among the Korean Native and Alien Horses Estimated by Microsatellite Polymorphism
 784 Genetic Relationship among the Korean Native and Alien Horses Estimated by Microsatellite Polymorphism G. J. Cho* College of Veterinary Medicine, Kyungpook National University, Daegu 702-701, Korea
784 Genetic Relationship among the Korean Native and Alien Horses Estimated by Microsatellite Polymorphism G. J. Cho* College of Veterinary Medicine, Kyungpook National University, Daegu 702-701, Korea
mtdna Diversity and Origin of Chinese Mongolian Horses*
 1696 Asian-Aust. J. Anim. Sci. Vol. 21, No. 12 : 1696-1702 December 2008 www.ajas.info mtdna Diversity and Origin of Chinese Mongolian Horses* Jinlian Li, Youfei Shi 1, Caiyun Fan 2 and Dugarjaviin Manglai**
1696 Asian-Aust. J. Anim. Sci. Vol. 21, No. 12 : 1696-1702 December 2008 www.ajas.info mtdna Diversity and Origin of Chinese Mongolian Horses* Jinlian Li, Youfei Shi 1, Caiyun Fan 2 and Dugarjaviin Manglai**
Mitochondrial DNA sequence diversity in extant Irish horse populations and in ancient horses
 SHORT COMMUNICATION doi:10.1111/j.1365-2052.2006.01506.x Mitochondrial DNA sequence diversity in extant Irish horse populations and in ancient horses A. M. McGahern*, C. J. Edwards, M. A. Bower, A. Heffernan,,
SHORT COMMUNICATION doi:10.1111/j.1365-2052.2006.01506.x Mitochondrial DNA sequence diversity in extant Irish horse populations and in ancient horses A. M. McGahern*, C. J. Edwards, M. A. Bower, A. Heffernan,,
History of Lipizzan horse maternal lines as revealed by mtdna analysis
 History of Lipizzan horse maternal lines as revealed by mtdna analysis Tatjana Kavar, Gottfried Brem, Franc Habe, Johann Sölkner, Peter Dovč To cite this version: Tatjana Kavar, Gottfried Brem, Franc Habe,
History of Lipizzan horse maternal lines as revealed by mtdna analysis Tatjana Kavar, Gottfried Brem, Franc Habe, Johann Sölkner, Peter Dovč To cite this version: Tatjana Kavar, Gottfried Brem, Franc Habe,
MOLECULAR PHYLOGENETIC RELATIOSHIPS IN ROMANIAN CYPRINIDS BASED ON cox1 AND cox2 SEQUENCES
 PROCEEDINGS OF THE BALKAN SCIENTIFIC CONFERENCE OF BIOLOGY IN PLOVDIV (BULGARIA) FROM 19 TH TILL 21 ST OF MAY 2005 (EDS B. GRUEV, M. NIKOLOVA AND A. DONEV), 2005 (P. 162 167) MOLECULAR PHYLOGENETIC RELATIOSHIPS
PROCEEDINGS OF THE BALKAN SCIENTIFIC CONFERENCE OF BIOLOGY IN PLOVDIV (BULGARIA) FROM 19 TH TILL 21 ST OF MAY 2005 (EDS B. GRUEV, M. NIKOLOVA AND A. DONEV), 2005 (P. 162 167) MOLECULAR PHYLOGENETIC RELATIOSHIPS
Genetic analysis of radio-tagged westslope cutthroat trout from St. Mary s River and Elk River. April 9, 2002
 Genetic analysis of radio-tagged westslope cutthroat trout from St. Mary s River and Elk River April 9, 2002 Report prepared for: Angela Prince, M.Sc., R.P. Bio Westslope Fisheries 517 13 th Avenue South
Genetic analysis of radio-tagged westslope cutthroat trout from St. Mary s River and Elk River April 9, 2002 Report prepared for: Angela Prince, M.Sc., R.P. Bio Westslope Fisheries 517 13 th Avenue South
Iberian Origins of New World Horse Breeds
 Journal of Heredity 2006:97(2):107 113 doi:10.1093/jhered/esj020 Advance Access publication February 17, 2006 Iberian Origins of New World Horse Breeds CRISTINA LUÍS, CRISTIANE BASTOS-SILVEIRA, E. GUS
Journal of Heredity 2006:97(2):107 113 doi:10.1093/jhered/esj020 Advance Access publication February 17, 2006 Iberian Origins of New World Horse Breeds CRISTINA LUÍS, CRISTIANE BASTOS-SILVEIRA, E. GUS
Characterization of two microsatellite PCR multiplexes for high throughput. genotyping of the Caribbean spiny lobster, Panulirus argus
 Characterization of two microsatellite PCR multiplexes for high throughput genotyping of the Caribbean spiny lobster, Panulirus argus Nathan K. Truelove 1, Richard F. Preziosi 1, Donald Behringer Jr 2,
Characterization of two microsatellite PCR multiplexes for high throughput genotyping of the Caribbean spiny lobster, Panulirus argus Nathan K. Truelove 1, Richard F. Preziosi 1, Donald Behringer Jr 2,
Quick method for identifying horse (Equus caballus) and donkey (Equus asinus) hybrids
 Short Communication Quick method for identifying horse (Equus caballus) and donkey (Equus asinus) hybrids M.M. Franco 1,2,3, J.B.F. Santos 2, A.S. Mendonça 3, T.C.F. Silva 2, R.C. Antunes 2 and E.O. Melo
Short Communication Quick method for identifying horse (Equus caballus) and donkey (Equus asinus) hybrids M.M. Franco 1,2,3, J.B.F. Santos 2, A.S. Mendonça 3, T.C.F. Silva 2, R.C. Antunes 2 and E.O. Melo
A pedigree-based study of mitochondrial D-loop DNA sequence variation among Arabian horses
 Animal Genetics, 2000, 31, 1±7 A pedigree-based study of mitochondrial D-loop DNA sequence variation among Arabian horses A T Bowling, A Del Valle, M Bowling A T Bowling A Del Valle M Bowling Veterinary
Animal Genetics, 2000, 31, 1±7 A pedigree-based study of mitochondrial D-loop DNA sequence variation among Arabian horses A T Bowling, A Del Valle, M Bowling A T Bowling A Del Valle M Bowling Veterinary
Continued Genetic Monitoring of the Kootenai Tribe of Idaho White Sturgeon Conservation Aquaculture Program
 Continued Genetic Monitoring of the Kootenai Tribe of Idaho White Sturgeon Conservation Aquaculture Program Deliverable 1): Monitoring of Kootenai River white sturgeon genetic diversity Deliverable 2):
Continued Genetic Monitoring of the Kootenai Tribe of Idaho White Sturgeon Conservation Aquaculture Program Deliverable 1): Monitoring of Kootenai River white sturgeon genetic diversity Deliverable 2):
59 th EAAP Annual Meeting August 24-27, 2008, Vilnius, Lithuania Abstract # 2908, Session 08
 59 th EAAP Annual Meeting August 24-27, 2008, Vilnius, Lithuania Abstract # 2908, Session 08 Genetic variability of the Skyros pony breed and its relationship with other Greek and foreign horse breeds
59 th EAAP Annual Meeting August 24-27, 2008, Vilnius, Lithuania Abstract # 2908, Session 08 Genetic variability of the Skyros pony breed and its relationship with other Greek and foreign horse breeds
wi Astuti, Hidayat Ashari, and Siti N. Prijono
 Phylogenetic position of Psittacula parakeet bird from Enggano Island, Indonesia based on analyses of cytochrome b gene sequences. wi Astuti, Hidayat Ashari, and Siti N. Prijono Research Centre for Biology,
Phylogenetic position of Psittacula parakeet bird from Enggano Island, Indonesia based on analyses of cytochrome b gene sequences. wi Astuti, Hidayat Ashari, and Siti N. Prijono Research Centre for Biology,
Initial microsatellite analysis of wild Kootenai River white sturgeon and subset brood stock groups used in a conservation aquaculture program.
 Initial microsatellite analysis of wild Kootenai River white sturgeon and subset brood stock groups used in a conservation aquaculture program. Prepared for: U.S. Department of Energy, Bonneville Power
Initial microsatellite analysis of wild Kootenai River white sturgeon and subset brood stock groups used in a conservation aquaculture program. Prepared for: U.S. Department of Energy, Bonneville Power
Genetic Analysis of the Pryor Mountains HMA, MT. E. Gus Cothran. September 2, 2010
 Genetic Analysis of the Pryor Mountains HMA, MT E. Gus Cothran September 2, 2010 Department of Veterinary Integrative Bioscience Texas A&M University College Station, TX 77843-4458 1 The following is a
Genetic Analysis of the Pryor Mountains HMA, MT E. Gus Cothran September 2, 2010 Department of Veterinary Integrative Bioscience Texas A&M University College Station, TX 77843-4458 1 The following is a
Assessment of Generation Interval and Inbreeding in Peruvian Paso Horse V. Montenegro 1,J. Vilela, 2 &M. Wurzinger 3
 Proceedings of the World Congress on Genetics Applied to Livestock Production, 11.749 Assessment of Generation Interval and Inbreeding in Peruvian Paso Horse V. Montenegro 1,J. Vilela, 2 &M. Wurzinger
Proceedings of the World Congress on Genetics Applied to Livestock Production, 11.749 Assessment of Generation Interval and Inbreeding in Peruvian Paso Horse V. Montenegro 1,J. Vilela, 2 &M. Wurzinger
Human Ancestry (Learning Objectives)
 Human Ancestry (Learning Objectives) 1. Identify the characters shared by all primates and relate them to the function they served in their common ancestor. 2. Learn the fields study of Human evolution
Human Ancestry (Learning Objectives) 1. Identify the characters shared by all primates and relate them to the function they served in their common ancestor. 2. Learn the fields study of Human evolution
Hybridization versus Randomly-Sorting Ancestral Alleles: Genetic Variation in Lake Malawi Cichlids
 Hybridization versus Randomly-Sorting Ancestral Alleles: Genetic Variation in Lake Malawi Cichlids Meryl Mims Faculty Advisor: Professor Todd Streelman Undergraduate Honors Thesis, Spring 2007 Georgia
Hybridization versus Randomly-Sorting Ancestral Alleles: Genetic Variation in Lake Malawi Cichlids Meryl Mims Faculty Advisor: Professor Todd Streelman Undergraduate Honors Thesis, Spring 2007 Georgia
CHAPTER III RESULTS. sampled from 22 streams, representing 4 major river drainages in New Jersey, and 1 trout
 CHAPTER III RESULTS Genetic Diversity Genotypes at 13 microsatellite DNA loci were determined for 238 brook trout sampled from 22 streams, representing 4 major river drainages in New Jersey, and 1 trout
CHAPTER III RESULTS Genetic Diversity Genotypes at 13 microsatellite DNA loci were determined for 238 brook trout sampled from 22 streams, representing 4 major river drainages in New Jersey, and 1 trout
georgii (TELEOSTEI: ISTIOPHORIDAE):
 BULLETIN OF MARINE SCIENCE, 79(3): 483 491, 2006 Validity, Identification, and Distribution of the Roundscale Spearfish, Tetrapturus georgii (TELEOSTEI: ISTIOPHORIDAE): Morphological and Molecular evidence
BULLETIN OF MARINE SCIENCE, 79(3): 483 491, 2006 Validity, Identification, and Distribution of the Roundscale Spearfish, Tetrapturus georgii (TELEOSTEI: ISTIOPHORIDAE): Morphological and Molecular evidence
57th Annual EAAP Meeting, Antalya, September
 G3.10 corresponding author: pariset@unitus.it @unitus.it. Genetic characterization by single nucleotide polymorphisms (SNPs) of Hungarian Grey and Maremmana cattle breeds L. Pariset 1, C. Marchitelli 1,
G3.10 corresponding author: pariset@unitus.it @unitus.it. Genetic characterization by single nucleotide polymorphisms (SNPs) of Hungarian Grey and Maremmana cattle breeds L. Pariset 1, C. Marchitelli 1,
SUMMARY KEYWORDS. Bluefin tuna, Population Genetics, Electronic Tagging, Spawning Migrations
 SCRS/2006/089 GENETIC DATA AND ELECTRONIC TAGGING INDICATE THAT THE GULF OF MEXICO AND MEDITERRANEAN SEA ARE REPRODUCTIVELY ISOLATED STOCKS OF BLUEFIN TUNA (THUNNUS THYNNUS) Andre M. Boustany 1, Carol
SCRS/2006/089 GENETIC DATA AND ELECTRONIC TAGGING INDICATE THAT THE GULF OF MEXICO AND MEDITERRANEAN SEA ARE REPRODUCTIVELY ISOLATED STOCKS OF BLUEFIN TUNA (THUNNUS THYNNUS) Andre M. Boustany 1, Carol
Hierarchical analysis of genetic structure in Spanish donkey breeds using microsatellite markers
 (2002) 89, 207 211 2002 Nature Publishing Group All rights reserved 0018-067X/02 $25.00 www.nature.com/hdy Hierarchical analysis of genetic structure in Spanish donkey breeds using microsatellite markers
(2002) 89, 207 211 2002 Nature Publishing Group All rights reserved 0018-067X/02 $25.00 www.nature.com/hdy Hierarchical analysis of genetic structure in Spanish donkey breeds using microsatellite markers
Assigning breed origin to alleles in crossbred animals
 DOI 10.1186/s12711-016-0240-y Genetics Selection Evolution RESEARCH ARTICLE Open Access Assigning breed origin to alleles in crossbred animals Jérémie Vandenplas 1*, Mario P. L. Calus 1, Claudia A. Sevillano
DOI 10.1186/s12711-016-0240-y Genetics Selection Evolution RESEARCH ARTICLE Open Access Assigning breed origin to alleles in crossbred animals Jérémie Vandenplas 1*, Mario P. L. Calus 1, Claudia A. Sevillano
Maternal phylogenetic relationships and genetic variation among Arabian horse populations using whole mitochondrial DNA D-loop sequencing
 Khanshour and Cothran BMC Genetics 2013, 14:83 RESEARCH ARTICLE Open Access Maternal phylogenetic relationships and genetic variation among Arabian horse populations using whole mitochondrial DNA D-loop
Khanshour and Cothran BMC Genetics 2013, 14:83 RESEARCH ARTICLE Open Access Maternal phylogenetic relationships and genetic variation among Arabian horse populations using whole mitochondrial DNA D-loop
Revealing the Past and Present of Bison Using Genome Analysis
 Revealing the Past and Present of Bison Using Genome Analysis David Forgacs, Rick Wallen, Lauren Dobson, Amy Boedeker, and James Derr July 5, 2017 1 Presentation outline 1. What can genetics teach us about
Revealing the Past and Present of Bison Using Genome Analysis David Forgacs, Rick Wallen, Lauren Dobson, Amy Boedeker, and James Derr July 5, 2017 1 Presentation outline 1. What can genetics teach us about
BIODIVERSITY OF LAKE VICTORIA:
 BIODIVERSITY OF LAKE VICTORIA:.. ITS CONSERVATION AND SUSTAINABLE USE [THE UGANDAN VERSION] Ogutu-Ohwayo, R. and Ndawula L. National Agricultural Research Organisation, Fisheries Resources Research Institute,
BIODIVERSITY OF LAKE VICTORIA:.. ITS CONSERVATION AND SUSTAINABLE USE [THE UGANDAN VERSION] Ogutu-Ohwayo, R. and Ndawula L. National Agricultural Research Organisation, Fisheries Resources Research Institute,
Species concepts What are species?
 What are species? 1 and definitions continue to be one of the most controversial topics in biology. A large number of alternative species concepts exist, each with its own strengths and limitations. Typological
What are species? 1 and definitions continue to be one of the most controversial topics in biology. A large number of alternative species concepts exist, each with its own strengths and limitations. Typological
Neglected Taxonomy of Rare Desert Fishes: Congruent Evidence for Two Species of Leatherside Chub
 Syst. Biol. 53(6):841 855, 2004 Copyright c Society of Systematic Biologists ISSN: 1063-5157 print / 1076-836X online DOI: 10.1080/10635150490522557 Neglected Taxonomy of Rare Desert Fishes: Congruent
Syst. Biol. 53(6):841 855, 2004 Copyright c Society of Systematic Biologists ISSN: 1063-5157 print / 1076-836X online DOI: 10.1080/10635150490522557 Neglected Taxonomy of Rare Desert Fishes: Congruent
Mitochondrial DNA analysis as a tool for family and species identification of fish larvae: Emphasis on Snappers.
 Mitochondrial DNA analysis as a tool for family and species identification of fish larvae: Emphasis on Snappers. Áurea E. Rodríguez, Juan C. Martínez-Cruzado, Ernesto Otero, Jorge R. García-Sais and Jennie
Mitochondrial DNA analysis as a tool for family and species identification of fish larvae: Emphasis on Snappers. Áurea E. Rodríguez, Juan C. Martínez-Cruzado, Ernesto Otero, Jorge R. García-Sais and Jennie
SHORT COMMUNICATION: Mitochondrial DNA D-loop sequence diversity and origin of Chinese pony breeds (Equus caballus)
 SHORT COMMUNICATION: Mitochondrial DNA D-loop sequence diversity and origin of Chinese pony breeds (Equus caballus) Sheng-lin Yang 1, Ai-Ping Li 2, Long-xing Xu 3, and Haibing Yang 1 1 Key Laboratory of
SHORT COMMUNICATION: Mitochondrial DNA D-loop sequence diversity and origin of Chinese pony breeds (Equus caballus) Sheng-lin Yang 1, Ai-Ping Li 2, Long-xing Xu 3, and Haibing Yang 1 1 Key Laboratory of
Genetic analyses of carp, goldfish, and carp goldfish hybrids in New Zealand
 Genetic analyses of carp, goldfish, and carp goldfish hybrids in New Zealand DOC RESEARCH & DEVELOPMENT SERIES 219 P.J. Smith and S.M. McVeagh Published by Science & Technical Publishing Department of
Genetic analyses of carp, goldfish, and carp goldfish hybrids in New Zealand DOC RESEARCH & DEVELOPMENT SERIES 219 P.J. Smith and S.M. McVeagh Published by Science & Technical Publishing Department of
MOLECULAR CHARACTERISATION AND PHYLOGENETICS OF MALAYSIAN GREEN AROWANA (Scleropages formosus) IN PENINSULAR MALAYSIA
 First ASIAHORCs Joint Symposium 18-20 July 2009 Nagoya, Japan MOLECULAR CHARACTERISATION AND PHYLOGENETICS OF MALAYSIAN GREEN AROWANA (Scleropages formosus) IN PENINSULAR MALAYSIA M. Rizman-Idid 1, S.
First ASIAHORCs Joint Symposium 18-20 July 2009 Nagoya, Japan MOLECULAR CHARACTERISATION AND PHYLOGENETICS OF MALAYSIAN GREEN AROWANA (Scleropages formosus) IN PENINSULAR MALAYSIA M. Rizman-Idid 1, S.
AN ISSUE OF GENETIC INTEGRITY AND DIVERSITY: ASSESSING THE CONSERVATION VALUE OF A PRIVATE AMERICAN BISON HERD
 AN ISSUE OF GENETIC INTEGRITY AND DIVERSITY: ASSESSING THE CONSERVATION VALUE OF A PRIVATE AMERICAN BISON HERD A Senior Scholars Thesis by ASHLEY SUZANNE MARSHALL Submitted to the Office of Undergraduate
AN ISSUE OF GENETIC INTEGRITY AND DIVERSITY: ASSESSING THE CONSERVATION VALUE OF A PRIVATE AMERICAN BISON HERD A Senior Scholars Thesis by ASHLEY SUZANNE MARSHALL Submitted to the Office of Undergraduate
Bighorn Sheep Research Activity Love Stowell & Ernest_1May2017 Wildlife Genomics & Disease Ecology Lab Updated 04/27/2017 SMLS
 Bighorn Sheep Research Activity 2016-17 Love Stowell & Ernest_1May2017 Wildlife Genomics & Disease Ecology Lab Updated 04/27/2017 SMLS Sample acquisition Samples acquired to date from sheep captured or
Bighorn Sheep Research Activity 2016-17 Love Stowell & Ernest_1May2017 Wildlife Genomics & Disease Ecology Lab Updated 04/27/2017 SMLS Sample acquisition Samples acquired to date from sheep captured or
SC China s Annual report Part II: The Squid Jigging Fishery Gang Li, Xinjun Chen and Bilin Liu
 3 rd Meeting of the Scientific Committee Port Vila, Vanuatu 28 September - 3 October 215 SC-3-9 China s Annual report Part II: The Squid Jigging Fishery Gang Li, Xinjun Chen and Bilin Liu National Report
3 rd Meeting of the Scientific Committee Port Vila, Vanuatu 28 September - 3 October 215 SC-3-9 China s Annual report Part II: The Squid Jigging Fishery Gang Li, Xinjun Chen and Bilin Liu National Report
An Update on Bison Genetics and Genomics"
 An Update on Bison Genetics and Genomics" Dr. James Derr, Professor Texas A&M University Texas Bison Association 26 October 2013 Crockett, Texas American Bison are found in all shapes, sizes and colors
An Update on Bison Genetics and Genomics" Dr. James Derr, Professor Texas A&M University Texas Bison Association 26 October 2013 Crockett, Texas American Bison are found in all shapes, sizes and colors
Evaluating genetic connectivity and re-colonization dynamics of moose in the Northeast.
 Evaluating genetic connectivity and re-colonization dynamics of moose in the Northeast. Theme 4, Principal Investigator(s): Dr. Heidi Kretser and Dr. Michale Glennon, Wildlife Conservation Society hkretser@wcs.org
Evaluating genetic connectivity and re-colonization dynamics of moose in the Northeast. Theme 4, Principal Investigator(s): Dr. Heidi Kretser and Dr. Michale Glennon, Wildlife Conservation Society hkretser@wcs.org
Research & Reviews: Journal of Veterinary Sciences
 Research & Reviews: Journal of Veterinary Sciences Genetic Diversity of Arabian Horse from Stud "Borike" (Bosnia and Herzegovina) Using Microsatellite Markers Dunja Rukavina 1 *, Danica Hasanbašić 1, Adaleta
Research & Reviews: Journal of Veterinary Sciences Genetic Diversity of Arabian Horse from Stud "Borike" (Bosnia and Herzegovina) Using Microsatellite Markers Dunja Rukavina 1 *, Danica Hasanbašić 1, Adaleta
Saving feral horse populations: does it really matter? A case study of wild horses from Doñ ana National Park in southern Spain
 Saving feral horse populations: does it really matter? A case study of wild horses from Doñ ana National Park in southern Spain J. L. Vega-Pla *, J. Calderó n, P. P. Rodríguez-Gallardo *, A. M. Martinez
Saving feral horse populations: does it really matter? A case study of wild horses from Doñ ana National Park in southern Spain J. L. Vega-Pla *, J. Calderó n, P. P. Rodríguez-Gallardo *, A. M. Martinez
Supplementary Materials for
 www.sciencemag.org/cgi/content/full/341/6149/847/dc1 Supplementary Materials for A Critical Crossroad for BLM s Wild Horse Program Robert A. Garrott* and Madan K. Oli This PDF file includes Materials and
www.sciencemag.org/cgi/content/full/341/6149/847/dc1 Supplementary Materials for A Critical Crossroad for BLM s Wild Horse Program Robert A. Garrott* and Madan K. Oli This PDF file includes Materials and
Rabbit MSTN gene polymorphisms and genetic effect analysis
 Rabbit MSTN gene polymorphisms and genetic effect analysis X.B. Qiao 1, K.Y. Xu 2, B. Li 1, X. Luan 1, T. Xia 1 and X.Z. Fan 1 1 College of Animal Science and Technology, Shandong Agricultural University,
Rabbit MSTN gene polymorphisms and genetic effect analysis X.B. Qiao 1, K.Y. Xu 2, B. Li 1, X. Luan 1, T. Xia 1 and X.Z. Fan 1 1 College of Animal Science and Technology, Shandong Agricultural University,
MALAWI CICHLIDS SARAH ROBBINS BSCI462 SPRING 2013
 MALAWI CICHLIDS SARAH ROBBINS BSCI462 SPRING 2013 CICHLIDS Family of fish within the infraclass Teleostei Over 1600 species discovered, Up to 3000 species predicted Most commonly found in Africa and South
MALAWI CICHLIDS SARAH ROBBINS BSCI462 SPRING 2013 CICHLIDS Family of fish within the infraclass Teleostei Over 1600 species discovered, Up to 3000 species predicted Most commonly found in Africa and South
Genetic Diversity of Chinese Indigenous Pig Breeds in Shandong Province Using Microsatellite Markers*
 28 Asian-Aust. J. Anim. Sci. Vol. 24, No. 1 : 28-36 January 2011 www.ajas.info Genetic Diversity of Chinese Indigenous Pig Breeds in Shandong Province Using Microsatellite Markers* J. Y. Wang 1,2, J. F.
28 Asian-Aust. J. Anim. Sci. Vol. 24, No. 1 : 28-36 January 2011 www.ajas.info Genetic Diversity of Chinese Indigenous Pig Breeds in Shandong Province Using Microsatellite Markers* J. Y. Wang 1,2, J. F.
JENJIT KHUDAMRONGSAWAT 1*, TUCKSAORN BHUMMAKASIKARA 2 AND NANTARIKA CHANSUE 3
 Tropical Natural History 17(1): 175-180, April 2017 2017 by Chulalongkorn University Short Note Preliminary Study of Genetic Diversity in the Giant Freshwater Stingray, Himantura chaophraya (Batoidea:
Tropical Natural History 17(1): 175-180, April 2017 2017 by Chulalongkorn University Short Note Preliminary Study of Genetic Diversity in the Giant Freshwater Stingray, Himantura chaophraya (Batoidea:
The importance of Pedigree in Livestock Breeding. Libby Henson and Grassroots Systems Ltd
 The importance of Pedigree in Livestock Breeding Libby Henson and Grassroots Systems Ltd Spotted sticks and Spotted rams Control Natural Selection Winter food supply Shelter Predation Disease control Impose
The importance of Pedigree in Livestock Breeding Libby Henson and Grassroots Systems Ltd Spotted sticks and Spotted rams Control Natural Selection Winter food supply Shelter Predation Disease control Impose
Lecture 2 Phylogenetics of Fishes. 1. Phylogenetic systematics. 2. General fish evolution. 3. Molecular systematics & Genetic approaches
 Lecture 2 Phylogenetics of Fishes 1. Phylogenetic systematics 2. General fish evolution 3. Molecular systematics & Genetic approaches Charles Darwin & Alfred Russel Wallace All species are related through
Lecture 2 Phylogenetics of Fishes 1. Phylogenetic systematics 2. General fish evolution 3. Molecular systematics & Genetic approaches Charles Darwin & Alfred Russel Wallace All species are related through
Salmon bycatch patterns in the Bering Sea pollock fishery
 Salmon bycatch patterns in the Bering Sea pollock fishery James Ianelli Seattle, WA Data from the North Pacific Observer Program (Fisheries Monitoring and Assessment) were analyzed for seasonal, temporal,
Salmon bycatch patterns in the Bering Sea pollock fishery James Ianelli Seattle, WA Data from the North Pacific Observer Program (Fisheries Monitoring and Assessment) were analyzed for seasonal, temporal,
Genetic Variations of Common Carp (Cyprinus carpio L.) In South-Eastern Part of Caspian Sea Using Five Microsatellite Loci
 World Journal of Zoology 6 (1): 56-60, 2011 ISSN 1817-3098 IDOSI Publications, 2011 Genetic Variations of Common Carp (Cyprinus carpio L.) In South-Eastern Part of Caspian Sea Using Five Microsatellite
World Journal of Zoology 6 (1): 56-60, 2011 ISSN 1817-3098 IDOSI Publications, 2011 Genetic Variations of Common Carp (Cyprinus carpio L.) In South-Eastern Part of Caspian Sea Using Five Microsatellite
Genetic characteristics of common carp (Cyprinus carpio) in Ireland
 Genetic characteristics of common carp (Cyprinus carpio) in Ireland Research by: Bill Brazier, Tom Cross, Eileen Dillane, Phil McGinnity, Simon Harrison, Debbie Chapman (UCC) & Jens Carlsson (UCD) Contents
Genetic characteristics of common carp (Cyprinus carpio) in Ireland Research by: Bill Brazier, Tom Cross, Eileen Dillane, Phil McGinnity, Simon Harrison, Debbie Chapman (UCC) & Jens Carlsson (UCD) Contents
Genetic variability and efficiency of DNA microsatellite markers for paternity testing in horse breeds from the Brazilian Marajó archipelago
 Short Communication Genetics and Molecular Biology, 31, 1, 68-72 (2008) Copyright by the Brazilian Society of Genetics. Printed in Brazil www.sbg.org.br Genetic variability and efficiency of DNA microsatellite
Short Communication Genetics and Molecular Biology, 31, 1, 68-72 (2008) Copyright by the Brazilian Society of Genetics. Printed in Brazil www.sbg.org.br Genetic variability and efficiency of DNA microsatellite
Molecular phylogeny of the Romanian cyprinids from the Danube River
 Roumanian Biotechnological Letters Vol. 13, No. 5, 2008, pp. 3970 3975 Copyright 2008 Bucharest University Printed in Romania. All rights reserved Roumanian Society of Biological Sciences ORIGINAL PAPER
Roumanian Biotechnological Letters Vol. 13, No. 5, 2008, pp. 3970 3975 Copyright 2008 Bucharest University Printed in Romania. All rights reserved Roumanian Society of Biological Sciences ORIGINAL PAPER
The Sustainability of Atlantic Salmon (Salmo salar L.) in South West England
 The Sustainability of Atlantic Salmon (Salmo salar L.) in South West England Submitted by Sarah-Louise Counter to the University of Exeter as a thesis for the degree of Doctor of Philosophy in Biological
The Sustainability of Atlantic Salmon (Salmo salar L.) in South West England Submitted by Sarah-Louise Counter to the University of Exeter as a thesis for the degree of Doctor of Philosophy in Biological
FOUNDER CONTRIBUTION IN THE ENDANGERED CZECH DRAUGHT HORSE BREEDS 1
 4 th Int. Symp. Animal Science Days, Ptuj, Slovenia, Sept. st 3 rd, 06. COBISS:.08 Agris category code: L0 FOUNDER CONTRIBUTION IN THE ENDANGERED CZECH DRAUGHT HORSE BREEDS Hana VOSTRÁ-VYDROVÁ, Luboš VOSTRÝ,
4 th Int. Symp. Animal Science Days, Ptuj, Slovenia, Sept. st 3 rd, 06. COBISS:.08 Agris category code: L0 FOUNDER CONTRIBUTION IN THE ENDANGERED CZECH DRAUGHT HORSE BREEDS Hana VOSTRÁ-VYDROVÁ, Luboš VOSTRÝ,
Anguilla marmorata (Giant Mottled Eel) Discovered in a New Location: Natural Range Expansion or Recent Human Introduction? 1
 Anguilla marmorata (Giant Mottled Eel) Discovered in a New Location: Natural Range Expansion or Recent Human Introduction? 1 Alex Handler 2 and Shelley A. James 3 Abstract: Freshwater eels in the family
Anguilla marmorata (Giant Mottled Eel) Discovered in a New Location: Natural Range Expansion or Recent Human Introduction? 1 Alex Handler 2 and Shelley A. James 3 Abstract: Freshwater eels in the family
UNIVERSITY OF ILLINOIS AT URBANA-CHAMPAIGN PRODUCTION NOTE. University of Illinois at Urbana-Champaign Library Large-scale Digitization Project, 2007.
 I L L IN 0 I S UNIVERSITY OF ILLINOIS AT URBANA-CHAMPAIGN PRODUCTION NOTE University of Illinois at Urbana-Champaign Library Large-scale Digitization Project, 2007. ci h oo /9 Illinois Natural History
I L L IN 0 I S UNIVERSITY OF ILLINOIS AT URBANA-CHAMPAIGN PRODUCTION NOTE University of Illinois at Urbana-Champaign Library Large-scale Digitization Project, 2007. ci h oo /9 Illinois Natural History
Supplemental figure 1. Collection sites and numbers of strains sequenced.
 Supplemental figure 1. Collection sites and numbers of strains sequenced. Supplemental figure 2. Neighbor joining 16S rrna gene phylogeny (636 bp). Minimum bootstrap values >50% generated using NJ and
Supplemental figure 1. Collection sites and numbers of strains sequenced. Supplemental figure 2. Neighbor joining 16S rrna gene phylogeny (636 bp). Minimum bootstrap values >50% generated using NJ and
PASTELARIA STUDIOS PUBLISHER
 PASTELARIA STUDIOS PUBLISHER WWW.PASTELARIASTUDIOS.COM Email pastelariastudios@gmail.com Teresa Maria Queiroz Pastelaria Studios New Release March 2015 capa ISBN 978-989-8796-18-9 Book 40,00 Pvp 14.50
PASTELARIA STUDIOS PUBLISHER WWW.PASTELARIASTUDIOS.COM Email pastelariastudios@gmail.com Teresa Maria Queiroz Pastelaria Studios New Release March 2015 capa ISBN 978-989-8796-18-9 Book 40,00 Pvp 14.50
Occasionally white fleeced Ryeland sheep will produce coloured fleeced lambs.
 Genetic Testing Service available for Members Occasionally white fleeced Ryeland sheep will produce coloured fleeced lambs. The Society offers members a genetic testing service, carried out by Cardiff
Genetic Testing Service available for Members Occasionally white fleeced Ryeland sheep will produce coloured fleeced lambs. The Society offers members a genetic testing service, carried out by Cardiff
AmpFlSTR Identifiler PCR Amplification Kit
 Application Note Human Identification AmpFlSTR Identifiler PCR Amplification Kit In Applied Biosystems continual efforts to improve the quality of our products, we have made some modifications to the manufacturing
Application Note Human Identification AmpFlSTR Identifiler PCR Amplification Kit In Applied Biosystems continual efforts to improve the quality of our products, we have made some modifications to the manufacturing
Population structure and genetic diversity of Bovec sheep from Slovenia preliminary results
 Population structure and genetic diversity of Bovec sheep from Slovenia preliminary results M. Simčič, M. Žan Lotrič, D. Bojkovski, E. Ciani, I. Medugorac Introduction Jezersko Solčava sheep Bovec sheep
Population structure and genetic diversity of Bovec sheep from Slovenia preliminary results M. Simčič, M. Žan Lotrič, D. Bojkovski, E. Ciani, I. Medugorac Introduction Jezersko Solčava sheep Bovec sheep
Department of Horse Breeding, Poznań University of Life Sciences, Wołyńska 33, Poznań, Poland
 Animal Science Papers and Reports vol. 31 (2013) no. 2, 159-164 Institute of Genetics and Animal Breeding, Jastrzębiec, Poland Genotyping of coat color genes (MC1R, ASIP, PMEL17 and MATP) polymorphisms
Animal Science Papers and Reports vol. 31 (2013) no. 2, 159-164 Institute of Genetics and Animal Breeding, Jastrzębiec, Poland Genotyping of coat color genes (MC1R, ASIP, PMEL17 and MATP) polymorphisms
Mitochondrial diversity and the origin of Iberian sheep
 Genet. Sel. Evol. 39 (2007) 91 103 91 c INRA, EDP Sciences, 2006 DOI: 10.1051/gse:2006034 Original article Mitochondrial diversity and the origin of Iberian sheep Susana PEDROSA a, Juan-José ARRANZ a,
Genet. Sel. Evol. 39 (2007) 91 103 91 c INRA, EDP Sciences, 2006 DOI: 10.1051/gse:2006034 Original article Mitochondrial diversity and the origin of Iberian sheep Susana PEDROSA a, Juan-José ARRANZ a,
GENETICS OF BRACHYGNATHISM IN PERUVIAN PASO HORSE
 Session 19 Horse genetics-behaviour traits GENETICS OF BRACHYGNATHISM IN PERUVIAN PASO HORSE Roberto Mantovani, Linda Giosmin, Enrico Sturaro, Alessio Cecchinato, and Luis Robin Wong Ponce (*) Department
Session 19 Horse genetics-behaviour traits GENETICS OF BRACHYGNATHISM IN PERUVIAN PASO HORSE Roberto Mantovani, Linda Giosmin, Enrico Sturaro, Alessio Cecchinato, and Luis Robin Wong Ponce (*) Department
Identification of Species-Diagnostic SNP Markers in Tilapias Using ddradseq
 Identification of Species-Diagnostic SNP Markers in Tilapias Using ddradseq Mochamad Syaifudin, Michael Bekaërt, John B Taggart, Gideon Hulata 1, Helena D Cotta 2, Jean-François Baroiller 2, David J Penman
Identification of Species-Diagnostic SNP Markers in Tilapias Using ddradseq Mochamad Syaifudin, Michael Bekaërt, John B Taggart, Gideon Hulata 1, Helena D Cotta 2, Jean-François Baroiller 2, David J Penman
What DNA tells us about Walleye (& other fish) in the Great Lakes
 What DNA tells us about Walleye (& other fish) in the Great Lakes Carol Stepien, Douglas Murphy, Rachel Lohner, & Jo Ann Banda Great Lakes Genetics Lab Lake Erie Center University of Toledo What do we
What DNA tells us about Walleye (& other fish) in the Great Lakes Carol Stepien, Douglas Murphy, Rachel Lohner, & Jo Ann Banda Great Lakes Genetics Lab Lake Erie Center University of Toledo What do we
Genetic consequences of stocking with hatchery strain brown trout: experiences from Denmark. Michael M. Hansen
 Genetic consequences of stocking with hatchery strain brown trout: experiences from Denmark Michael M. Hansen Agenda Brown trout stocking in Denmark Introgression of stocked trout into wild populations
Genetic consequences of stocking with hatchery strain brown trout: experiences from Denmark Michael M. Hansen Agenda Brown trout stocking in Denmark Introgression of stocked trout into wild populations
Evidence for Introgressive Hybridization of Captive Markhor (Capra falconeri) with Domestic Goat: Cautions for Reintroduction
 Biochem Genet (2008) 46:216 226 DOI 10.1007/s10528-008-9145-y Evidence for Introgressive Hybridization of Captive Markhor (Capra falconeri) with Domestic Goat: Cautions for Reintroduction Sabine E. Hammer
Biochem Genet (2008) 46:216 226 DOI 10.1007/s10528-008-9145-y Evidence for Introgressive Hybridization of Captive Markhor (Capra falconeri) with Domestic Goat: Cautions for Reintroduction Sabine E. Hammer
BRIEF COMMUNICATION Phylogenetic relationships within the genus Pimephales as inferred from ND4 and ND4L nucleotide sequences
 Journal of Fish Biology (2002) 61, 293 297 doi:10.1006/jfbi.2002.2002, available online at http://www.idealibrary.com on BRIEF COMMUNICATION Phylogenetic relationships within the genus Pimephales as inferred
Journal of Fish Biology (2002) 61, 293 297 doi:10.1006/jfbi.2002.2002, available online at http://www.idealibrary.com on BRIEF COMMUNICATION Phylogenetic relationships within the genus Pimephales as inferred
Little-known history of our native breeds reveals great sport horse genes.
 Little-known history of our native breeds reveals great sport horse genes. by Kathleen Kirsan For those of you who have been immersed in the Warmblood and sport horse culture of the last 30 years, I have
Little-known history of our native breeds reveals great sport horse genes. by Kathleen Kirsan For those of you who have been immersed in the Warmblood and sport horse culture of the last 30 years, I have
Farm Animals Breeding Act 1
 Issuer: Riigikogu Type: act In force from: 01.01.2015 In force until: 30.06.2017 Translation published: 10.04.2015 Farm Animals Breeding Act 1 Amended by the following acts Passed 06.11.2002 RT I 2002,
Issuer: Riigikogu Type: act In force from: 01.01.2015 In force until: 30.06.2017 Translation published: 10.04.2015 Farm Animals Breeding Act 1 Amended by the following acts Passed 06.11.2002 RT I 2002,
Genetic diversity in Spanish donkey breeds using microsatellite DNA markers
 Genet. Sel. Evol. 33 (2001) 433 442 433 INRA, EDP Sciences, 2001 Original article Genetic diversity in Spanish donkey breeds using microsatellite DNA markers José ARANGUREN-MÉNDEZ a,b, Jordi JORDANA a,,
Genet. Sel. Evol. 33 (2001) 433 442 433 INRA, EDP Sciences, 2001 Original article Genetic diversity in Spanish donkey breeds using microsatellite DNA markers José ARANGUREN-MÉNDEZ a,b, Jordi JORDANA a,,
Genetic diversity and population structure of Kazakh horses (Equus caballus) inferred from mtdna sequences
 Genetic diversity and population structure of Kazakh horses (Equus caballus) inferred from mtdna sequences M. Gemingguli 1, K.R. Iskhan 2, Y. Li 3, A. Qi 4, W. Wunirifu 5, L.Y. Ding 1 and A. Wumaierjiang
Genetic diversity and population structure of Kazakh horses (Equus caballus) inferred from mtdna sequences M. Gemingguli 1, K.R. Iskhan 2, Y. Li 3, A. Qi 4, W. Wunirifu 5, L.Y. Ding 1 and A. Wumaierjiang
Iou.. o MOLECULAR IEVOLUTION
 J Mol Evol (1995) 41:180-188 Iou.. o MOLECULAR IEVOLUTION Springer-Vedag New York Inc. 1995 Mitochondrial DNA Sequences of Various Species of the Genus Equus with Special Reference to the Phylogenetic
J Mol Evol (1995) 41:180-188 Iou.. o MOLECULAR IEVOLUTION Springer-Vedag New York Inc. 1995 Mitochondrial DNA Sequences of Various Species of the Genus Equus with Special Reference to the Phylogenetic
International Journal of Research in Zoology. Original Article
 Available online at http://www.urpjournals.com International Journal of Research in Zoology Universal Research Publications. All rights reserved Original Article ISSN 2278 1358 Phylogeny and genetic divergence
Available online at http://www.urpjournals.com International Journal of Research in Zoology Universal Research Publications. All rights reserved Original Article ISSN 2278 1358 Phylogeny and genetic divergence
A note on the population structure of leopards (Panthera pardus) in South Africa
 A note on the population structure of leopards (Panthera pardus) in South Africa Laura Tensen 1 *, Dick Roelofs 2 & Lourens H. Swanepoel 3,4 1 Department of Zoology, Faculty of Science, University of Johannesburg,
A note on the population structure of leopards (Panthera pardus) in South Africa Laura Tensen 1 *, Dick Roelofs 2 & Lourens H. Swanepoel 3,4 1 Department of Zoology, Faculty of Science, University of Johannesburg,
Employer Name: NOAA Fisheries, Northwest Fisheries Science Center
 Internship Description Employer Name: NOAA Fisheries, Northwest Fisheries Science Center Employer Description: Manchester Research Station functions as a satellite facility to the NOAA Fisheries Northwest
Internship Description Employer Name: NOAA Fisheries, Northwest Fisheries Science Center Employer Description: Manchester Research Station functions as a satellite facility to the NOAA Fisheries Northwest
Section 5 2 Limits To Growth Pages
 Section 5 2 Limits To Growth Pages 124 127 Free PDF ebook Download: Section 5 2 Limits To Growth Pages 124 127 Download or Read Online ebook section 5 2 limits to growth pages 124 127 in PDF Format From
Section 5 2 Limits To Growth Pages 124 127 Free PDF ebook Download: Section 5 2 Limits To Growth Pages 124 127 Download or Read Online ebook section 5 2 limits to growth pages 124 127 in PDF Format From
Legendre et al Appendices and Supplements, p. 1
 Legendre et al. 2010 Appendices and Supplements, p. 1 Appendices and Supplement to: Legendre, P., M. De Cáceres, and D. Borcard. 2010. Community surveys through space and time: testing the space-time interaction
Legendre et al. 2010 Appendices and Supplements, p. 1 Appendices and Supplement to: Legendre, P., M. De Cáceres, and D. Borcard. 2010. Community surveys through space and time: testing the space-time interaction
Matthew Alan Bertone
 Matthew Alan Bertone HOME: 109 Dunnsbee Drive - Garner, NC 27529 - phone: 919.210.9857 OFFICE: North Carolina State University - Campus Box 7613 - Raleigh, NC 27695 phone: 919.515.3429 - fax: 919.515.7746
Matthew Alan Bertone HOME: 109 Dunnsbee Drive - Garner, NC 27529 - phone: 919.210.9857 OFFICE: North Carolina State University - Campus Box 7613 - Raleigh, NC 27695 phone: 919.515.3429 - fax: 919.515.7746
DNA Breed Profile Testing FAQ. What is the history behind breed composition testing?
 DNA Breed Profile Testing FAQ What is the history behind breed composition testing? Breed composition testing, or breed integrity testing has been in existence for many years. Hampshire, Landrace, and
DNA Breed Profile Testing FAQ What is the history behind breed composition testing? Breed composition testing, or breed integrity testing has been in existence for many years. Hampshire, Landrace, and
IRISH NATIONAL STUD NICKING GUIDE
 Index Page Sire Selection Box 1 Dam Selection Box 2 Pedigree Display 3 Nicking Type 4 Pedigree Nicking Level 5 Inbreds Button 6 Nick Button 7 Nick Results Glossary 8 PDF & Nick Score 9 Inbreeding Filter
Index Page Sire Selection Box 1 Dam Selection Box 2 Pedigree Display 3 Nicking Type 4 Pedigree Nicking Level 5 Inbreds Button 6 Nick Button 7 Nick Results Glossary 8 PDF & Nick Score 9 Inbreeding Filter
Peopling of the Marianas: An mtdna perspective
 Peopling of the Marianas: An mtdna perspective 2 nd Marianas History Conference August 31, 2013 Miguel G. Vilar 1,3,5 Daniel E. Lynch 1,2,3 Chim W. Chan 1,2,3 Dana R. Santos 1,3 Ralph M. Garruto 2,3,4
Peopling of the Marianas: An mtdna perspective 2 nd Marianas History Conference August 31, 2013 Miguel G. Vilar 1,3,5 Daniel E. Lynch 1,2,3 Chim W. Chan 1,2,3 Dana R. Santos 1,3 Ralph M. Garruto 2,3,4
Inferring human population history from multiple genome sequences
 Inferring human population history from multiple genome sequences Stephan Schiffels Postdoctoral Fellow with Richard Durbin, Wellcome Trust Sanger Institute, Cambridge, UK From genome sequences to human
Inferring human population history from multiple genome sequences Stephan Schiffels Postdoctoral Fellow with Richard Durbin, Wellcome Trust Sanger Institute, Cambridge, UK From genome sequences to human
Phylogenetic analysis among Cyprinidae family using 16SrRNA
 2014; 1(6): 66-71 ISSN: 2347-5129 IJFAS 2014; 1(6): 66-71 2013 IJFAS www.fisheriesjournal.com Received: 20-05-2014 Accepted: 02-06-2014 Utpala Sharma Varsha singhal Dayal P. Gupta Partha Sarathi Mohanty
2014; 1(6): 66-71 ISSN: 2347-5129 IJFAS 2014; 1(6): 66-71 2013 IJFAS www.fisheriesjournal.com Received: 20-05-2014 Accepted: 02-06-2014 Utpala Sharma Varsha singhal Dayal P. Gupta Partha Sarathi Mohanty
Assessment of demographic bottleneck in Indian horse and endangered pony breeds
 c Indian Academy of Sciences ONLINE RESOURCES Assessment of demographic bottleneck in Indian horse and endangered pony breeds A. K. GUPTA 1, MAMTA CHAUHAN 1, ANURADHA BHARDWAJ 1 and R. K. VIJH 2 1 National
c Indian Academy of Sciences ONLINE RESOURCES Assessment of demographic bottleneck in Indian horse and endangered pony breeds A. K. GUPTA 1, MAMTA CHAUHAN 1, ANURADHA BHARDWAJ 1 and R. K. VIJH 2 1 National
Species Identification of small juvenile tunas caught in surface fisheries in the Phili... 1/13 ページ
 Species Identification of small juvenile tunas caught in surface fisheries in the Phili... 1/13 ページ Originated by: Fisheries and Aquaculture Department Title: Status of Interactions of Pacific Tuna Fisheries
Species Identification of small juvenile tunas caught in surface fisheries in the Phili... 1/13 ページ Originated by: Fisheries and Aquaculture Department Title: Status of Interactions of Pacific Tuna Fisheries
Analysis of Variance. Copyright 2014 Pearson Education, Inc.
 Analysis of Variance 12-1 Learning Outcomes Outcome 1. Understand the basic logic of analysis of variance. Outcome 2. Perform a hypothesis test for a single-factor design using analysis of variance manually
Analysis of Variance 12-1 Learning Outcomes Outcome 1. Understand the basic logic of analysis of variance. Outcome 2. Perform a hypothesis test for a single-factor design using analysis of variance manually
MANAGING WILDLIFE IN THE SOUTHWEST: New Challenges for the 21 st Century
 MANAGING WILDLIFE IN THE SOUTHWEST: New Challenges for the 21 st Century Proceedings of the Symposium August 2005 Edited by James W. Cain III and Paul R. Krausman A Publication of the Southwest Section
MANAGING WILDLIFE IN THE SOUTHWEST: New Challenges for the 21 st Century Proceedings of the Symposium August 2005 Edited by James W. Cain III and Paul R. Krausman A Publication of the Southwest Section
The genetic structure of Spanish Celtic horse breeds inferred from microsatellite data
 Animal Genetics, 2000, 31, 39±48 The genetic structure of Spanish Celtic horse breeds inferred from microsatellite data J CanÄ on, M L Checa, C Carleos, J L Vega-Pla, M Vallejo, S Dunner J CanÄ on M L
Animal Genetics, 2000, 31, 39±48 The genetic structure of Spanish Celtic horse breeds inferred from microsatellite data J CanÄ on, M L Checa, C Carleos, J L Vega-Pla, M Vallejo, S Dunner J CanÄ on M L
PCR primers for 100 microsatellites in red drum (Sciaenops ocellatus)
 Molecular Ecology Resources (2008) 8, 393 398 doi: 10.1111/j.1471-8286.2007.01969.x Blackwell Publishing Ltd PERMANENT GENETIC RESOURCES PCR primers for 100 microsatellites in red drum (Sciaenops ocellatus)
Molecular Ecology Resources (2008) 8, 393 398 doi: 10.1111/j.1471-8286.2007.01969.x Blackwell Publishing Ltd PERMANENT GENETIC RESOURCES PCR primers for 100 microsatellites in red drum (Sciaenops ocellatus)
Explore the basis for including competition traits in the genetic evaluation of the Icelandic horse
 Elsa Albertsdóttir The Swedish Agricultural University Faculty of Veterinary Medicine and Animal Science Department of Animal Breeding and Genetics Uppsala Explore the basis for including competition traits
Elsa Albertsdóttir The Swedish Agricultural University Faculty of Veterinary Medicine and Animal Science Department of Animal Breeding and Genetics Uppsala Explore the basis for including competition traits
16S rrna Gene Sequence Analysis of Snow Leopard, Gray Wolf, Horse and Bactrian Camel in Mongolia
 Journal of Agricultural Science and Technology A 7 (2017) 350-356 doi: 10.17265/2161-6256/2017.05.007 D DAVID PUBLISHING 16S rrna Gene Sequence Analysis of Snow Leopard, Gray Wolf, Horse Munkhtuul Tsogtgerel
Journal of Agricultural Science and Technology A 7 (2017) 350-356 doi: 10.17265/2161-6256/2017.05.007 D DAVID PUBLISHING 16S rrna Gene Sequence Analysis of Snow Leopard, Gray Wolf, Horse Munkhtuul Tsogtgerel
Diversity of Thermophilic Bacteria Isolated from Hot Springs
 ... 40(2) 524-533 (2555) KKU Sci. J. 40(2) 524-533 (2012) Diversity of Thermophilic Bacteria Isolated from Hot Springs ( 30, 3.65 x 10 5 1. x 10 3 CFU/ml (a, b, c, d, e f 16S rrna 1. kb Polymerase Chain
... 40(2) 524-533 (2555) KKU Sci. J. 40(2) 524-533 (2012) Diversity of Thermophilic Bacteria Isolated from Hot Springs ( 30, 3.65 x 10 5 1. x 10 3 CFU/ml (a, b, c, d, e f 16S rrna 1. kb Polymerase Chain
Advanced Animal Science TEKS/LINKS Student Objectives One Credit
 First Six Weeks Career/Safety/Work Habits AAS 1(A) The student will identify career development and entrepreneurship opportunities in the field of animal systems. AAS 1(B) The student will apply competencies
First Six Weeks Career/Safety/Work Habits AAS 1(A) The student will identify career development and entrepreneurship opportunities in the field of animal systems. AAS 1(B) The student will apply competencies
Level 3 Biology, 2011
 90719 907190 3SUPERVISOR S Level 3 Biology, 2011 90719 Describe trends in human evolution 2.00 pm uesday Tuesday 1 November 2011 Credits: Three Check that the National Student Number (NSN) on your admission
90719 907190 3SUPERVISOR S Level 3 Biology, 2011 90719 Describe trends in human evolution 2.00 pm uesday Tuesday 1 November 2011 Credits: Three Check that the National Student Number (NSN) on your admission
Parentage verification of Iranian Caspian horse using microsatellites markers
 Parentage verification of Iranian Caspian horse using microsatellites markers Hamidreza Seyedabadi *, Cyrus Amirinia, Mohammad Hossein Banabazi, Hossein Emrani Department of Biotechnology, Animal Science
Parentage verification of Iranian Caspian horse using microsatellites markers Hamidreza Seyedabadi *, Cyrus Amirinia, Mohammad Hossein Banabazi, Hossein Emrani Department of Biotechnology, Animal Science
Looking a fossil horse in the mouth! Using teeth to examine fossil horses!
 Looking a fossil horse in the mouth Using teeth to examine fossil horses Virginia Museum of Natural History Paleontology Department Fossil Teaching Kit 1 Teacher s Guide In this activity students will
Looking a fossil horse in the mouth Using teeth to examine fossil horses Virginia Museum of Natural History Paleontology Department Fossil Teaching Kit 1 Teacher s Guide In this activity students will
CONSERVATION OF LIVESTOCK BIODIVERSITY IN SPAIN
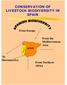 From Europe From the Mediterranean Area To Iberoamerica From Northern Africa EVOLUTION OF THE SPANISH GENETIC RESOURCES Until 18th and 19th Centuries: Traditional Systems: -Family Integrated management
From Europe From the Mediterranean Area To Iberoamerica From Northern Africa EVOLUTION OF THE SPANISH GENETIC RESOURCES Until 18th and 19th Centuries: Traditional Systems: -Family Integrated management
